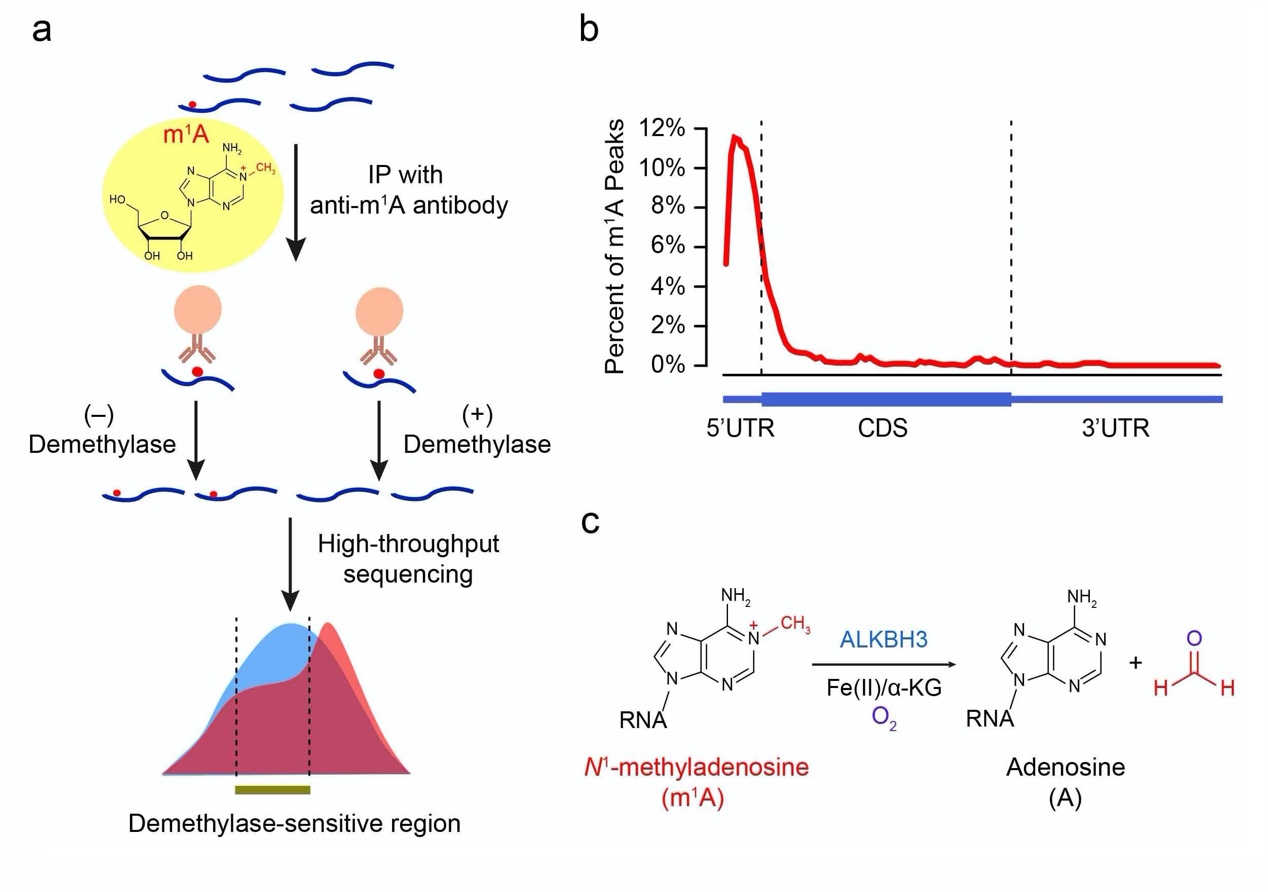
Our N1-methyladenosine methylome work is published in Nature Chemical Biology!
N1-Methyladenosine (m1A) is a prevalent post-transcriptional RNA modification, yet little is known about its abundance, topology and dynamics in mRNA. Here, we show that m1A is prevalent in Homo sapiens mRNA, which shows an m1A/A ratio of ~0.02%. We develop the m1A-ID-seq technique, based on m1A immunoprecipitation and the inherent ability of m1A to stall reverse transcription, as a means for transcriptome-wide m1A profiling. m1A-ID-seq identifies 901 m1A peaks (from 600 genes) in mRNA and noncoding RNA and reveals a prominent feature, enrichment in the 5’ untranslated region of mRNA transcripts, that is distinct from the pattern for N6-methyladenosine, the most abundant internal mammalian mRNA modification. Moreover, m1A in mRNA is reversible by ALKBH3, a known DNA/RNA demethylase. Lastly, we show that m1A methylation responds dynamically to stimuli, and we identify hundreds of stress-induced m1A sites. Collectively, our approaches allow comprehensive analysis of m1A dification and provide tools for functional studies of potential epigenetic regulation via the reversible and dynamic m1A methylation.