Our recent paper in Angewandte Chemie reports a sequencing method to detect cisplatin-DNA adducts in the human genome
Our new article named “Base-Resolution Analysis of Cisplatin–DNA Adducts at the Genome Scale” has been published in Angewandte Chemie as “Very Important Paper”(DOI : 10.1002/anie.201607380 in the International Edition;DOI : 10.1002/ange.201607380 in the German edition).
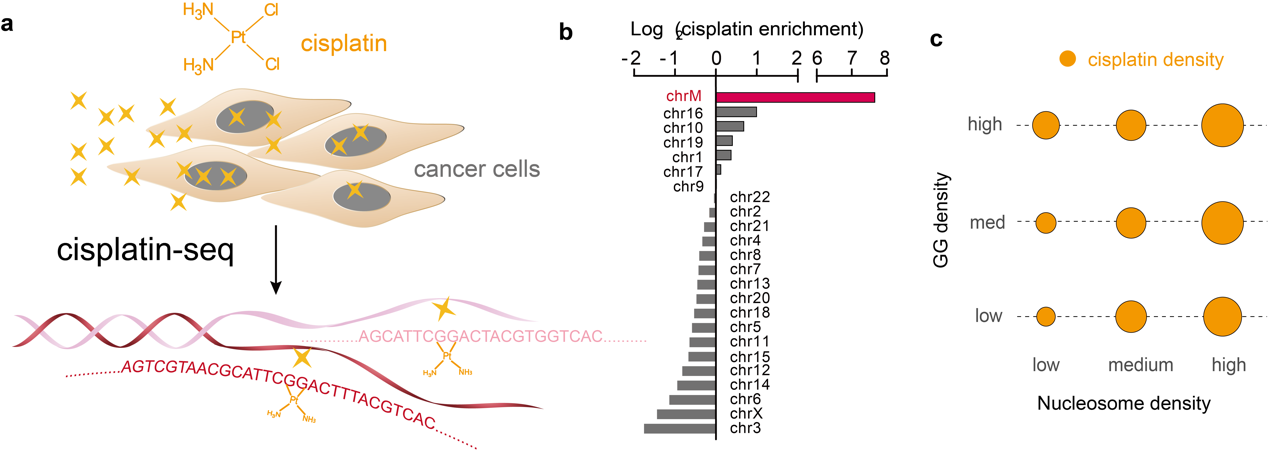
Cisplatin, one of the most widely used anticancer drugs, crosslinks DNA and ultimately induces cell death. However, the genomic pattern of cisplatin-DNA adducts remains unknown, due to the lack of a reliable and sensitive genome-wide method. Here we present “cisplatin-seq” to identify genome-wide cisplatin crosslinking sites at base-resolution. Cisplatin-seq reveals that mitochondrial DNA is a preferred target of cisplatin. For nuclear genome, cisplatin-DNA adducts are enriched within promoters and regions harboring transcription termination sites. While the density of GG dinucleotide determines the initial crosslinking of cisplatin, binding of proteins to the genome largely contributes to the accumulative pattern of cisplatin-DNA adducts.