We developed CLEVER-seq to detect 5-formylcytosine in pluripotent stem cells and mouse early embryos at single-cell and single-base resolution
Our new article named “Single-Cell 5-Formylcytosine Landscapes of Mammalian Early Embryos and ESCs at Single-Base Resolution” has been published in Cell Stem Cell (http://dx.doi.org/10.1016/j.stem.2017.02.013).
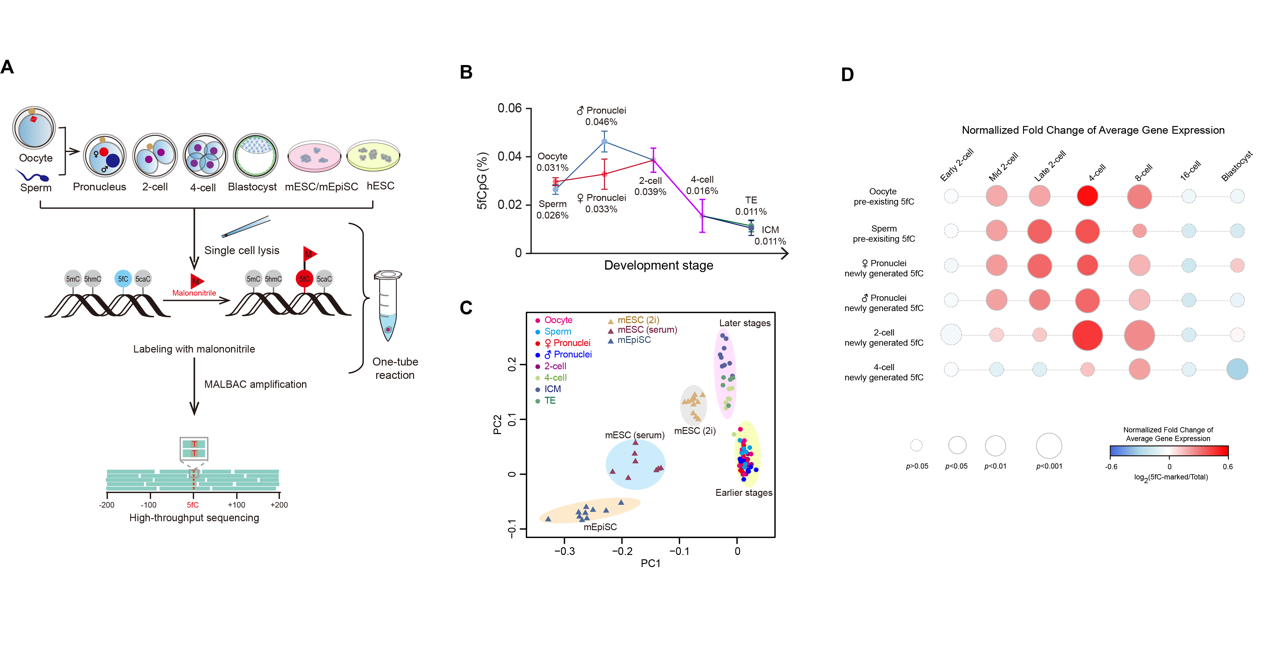
We present ‘‘CLEVER- seq’’ (chemical-labeling-enabled C-to-T conversion sequencing), which is a single-cell, single-base resolution 5fC-sequencing technology, based on biocompatible, selective chemical labeling of 5fC and subsequent C-to-T conversion during amplification and sequencing. CLEVER-seq shows intrinsic 5fC heterogeneity in mouse early embryos, Epi stem cells, and embryonic stem cells. CLEVER-seq of mouse early embryos also reveals the highly patterned genomic distribution and parental-specific dynamics of 5fC during mouse early pre-implantation development. Integrated analysis demonstrates that promoter 5fC production precedes the expression up-regulation of a clear set of developmentally and metabolically critical genes. Collectively, our work reveals the dynamics of active DNA demethylation during mouse pre-implantation development and provides an important resource for further functional studies of epigenetic reprogramming in single cells.