We revealed the mechanism of how unnatural cytosine bases recognized as Thymines by DNA polymerase
Our new article named “Unnatural Cytosine Bases Recognized as Thymines by DNA Poly- merases by the Formation of the Watson–Crick Geometry” has been published in Angewandte Chemie International Edition (https://doi.org/10.1002/anie.201807845 )
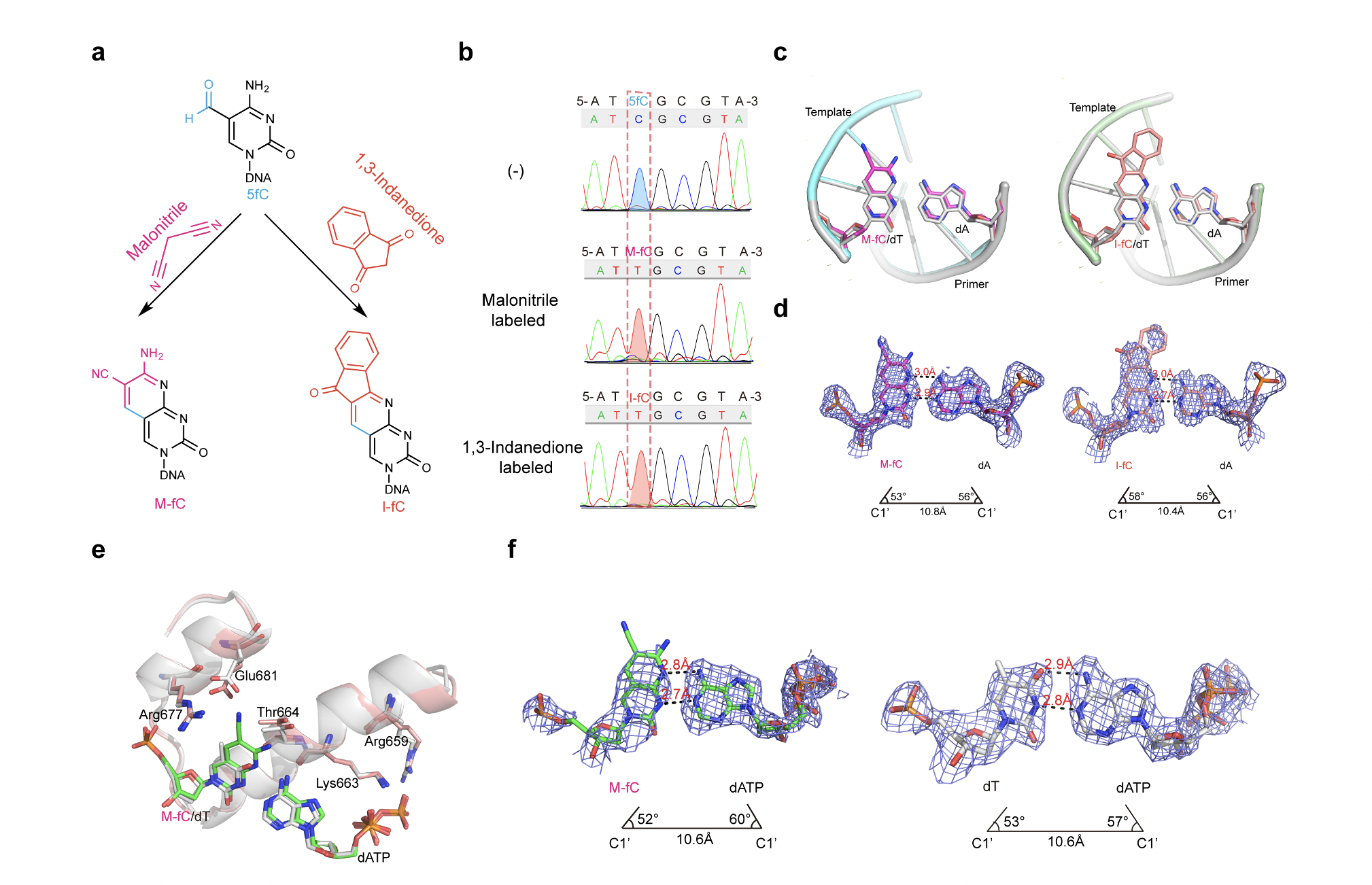
It was previously shown that two unnatural cytosine bases (termed “M‐fC” and “I‐fC”), which are chemical labeling adducts of the epigenetic base 5‐formylcytosine, can induce C‐to‐T transition during DNA amplification. However, how DNA polymerases recognize such unnatural cytosine bases remains enigmatic. To characterize the base pairing mode of the labeling adducts, we determined crystal structures of the labeling adducts pairing to dA/dG in the KlenTaq-host-guest complex system. The crystallographic observations suggest that both M-fC and I-fC can pair with an opposing dA via Watson– Crick geometry, in a position of DNA duplex devoid of contact with polymerase. We also solved the structures of the ternary complex in which M-fC base pairs with dATP at the incorporation site of KlenTaq polymerase. The results showed that the presence of the labeling adducts in the template strand is capable of inducing the formation of a Watson–Crick-like pair with an incoming dATP in the incorporation site of the DNA polymerase. Our study revealed that the formation of the Watson-Crick geometry was important for the recognition of the labeling adducts. This study expands the current understanding of unnatural base recognition by DNA polymerases.